DaPars2: Dynamics analysis of Alternative PolyAdenylation from RNA-seq Using Multiple Samples¶
Introduction¶
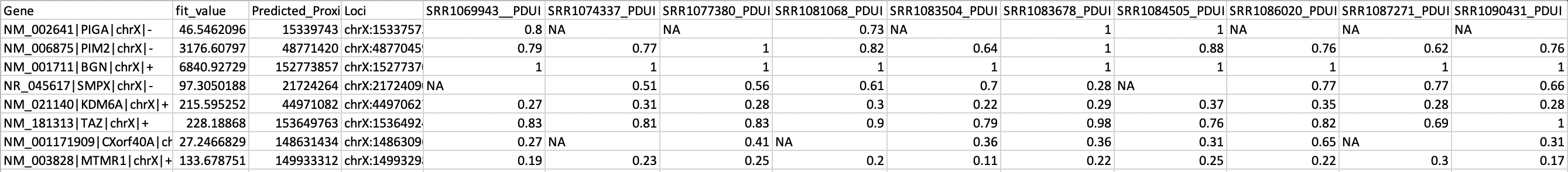
The dynamic usage of the 3’Untranslated Region (3’UTR) resulting from alternative polyadenylation (APA) is emerging as a pervasive mechanism for regulating mRNA diversity, stability and translation. Though RNA-seq provides the whole-transcriptome information and tools for analyzing gene/isoform expression are available, very few tool focus on the analysis of 3’UTR from standard RNA-seq. DaPars2 is the next generation of DaPars that directly infers the dynamic alternative polyadenylation (APA) usage by comparing standard RNA-seq from multiple samples. Given the annotated gene model, DaPars2 can infer the de novo proximal APA sites as well as the long and short 3’UTR expression levels. Finally, the dynamic APA usages of each samples will be identified.
Installation¶
Prerequisite: python2.7; numpy; scipy; R
Install DaPars2:
tar zxf DaPars2-VERSION.tar.gz
cd DaPars2-VERSION
Input format¶
DaPars2 requires the following two file formats as input:
Usage Information¶
Step 1: Generate region annotation: DaPars_Extract_Anno.py¶
DaPars2 will use the extracted distal polyadenylation sites to infer the proximal polyadenylation sites based on the alignment wiggle files of all samples. The output in this step will be used by step 3.
- Options:
-h, --help show this help message and exit -b GENE_BED_FILE, --bed=GENE_BED_FILE The gene model in BED format. The BED file can be downloaded from UCSC -s Gene_Symbol_FILE, --gene_symbol_map=Gene_Symbol_FILE The mapping of transcripts to gene symbol. -o OUTPUT_FILE, --out-prefix=OUTPUT_FILE The extracted annotation region will be stored into this file.
Run the following commands to generate region annotation analysis:
python DaPars_Extract_Anno.py -b hg19_refseq_whole_gene.bed -s hg19_4_19_2012_Refseq_id_from_UCSC.txt -o hg19_refseq_extracted_3UTR.bed
Step 2: Generate mapped reads files for all Samples¶
DaPars2 will use number of mapping reads for each samples to normalize library sizes. The output in this step will be used by step 3.
The format of the mapping reads file is:
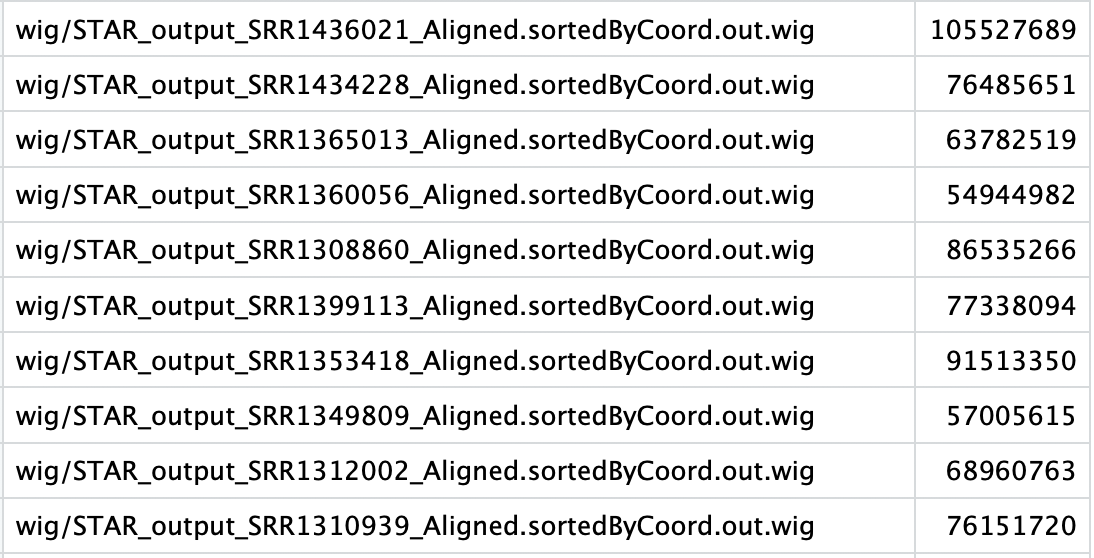
The first column is the sample name and the second column is the corresponding mapped reads. The order of the samples in the mapped reads file must be the same as the order of wiggles files in step 3.
Step 3: Run DaPars2: Dapars2_Multi_Sample.py ¶
Dapars2 uses multiple threads and analyzes each chromosome separately to save time and memory.
Run the following commands to perform DaPar2 analysis:
python Dapars2_Multi_Sample.py Dapars2_configure_file current_processing_chrmosome
The configure file contains most of the parameters for DaPars2_Multi_Sample.py.
The format of the configure is:
#The following file is the result of step 1.
Annotated_3UTR=hg19_refseq_extracted_3UTR.bed
#A comma-separated list of Wiggle files of all samples
Aligned_Wig_files=wig/STAR_output_SRR1436021_Aligned.sortedByCoord.out.wig,wig/STAR_output_SRR1434228_Aligned.sortedByCoord.out.wig,
wig/STAR_output_SRR1365013_Aligned.sortedByCoord.out.wig,wig/STAR_output_SRR1360056_Aligned.sortedByCoord.out.wig,
wig/STAR_output_SRR1308860_Aligned.sortedByCoord.out.wig,wig/STAR_output_SRR1399113_Aligned.sortedByCoord.out.wig,
wig/STAR_output_SRR1353418_Aligned.sortedByCoord.out.wig,wig/STAR_output_SRR1349809_Aligned.sortedByCoord.out.wig,
wig/STAR_output_SRR1312002_Aligned.sortedByCoord.out.wig,wig/STAR_output_SRR1310939_Aligned.sortedByCoord.out.wig
Output_directory=DaPars_Test_data/
Output_result_file=DaPars_Test_data
# Specify the Coverage_threshold
Coverage_threshold=10
# Specify the number of threads used to process the data
Num_Threads = 8
# Provide Sequencing_depth file for normalization
sequencing_depth_file=mapping_bam_location_with_depth.txt
Contact¶
- Lei Li: lei.li.bioinfo@gmail.com